Abstract
Acute myeloid leukemia (AML) is a heterogeneous disease characterized in part by genetic and epigenetic alterations. Cytarabine arabinoside (Ara-C) has been the cornerstone of chemotherapy treatment for patients diagnosed with AML for decades. Following cellular uptake, ara-C is phosphorylated into its active metabolite, ara-CTP, which primarily exerts its cytotoxic effects by inhibiting DNA synthesis in proliferating cells. Interpatient variation in the enzymes involved in ara-C activation/inactivation pathway (Fig 1A), can impact intracellular abundance of ara-CTP and thus its therapeutic benefit. Recently, deoxynucleoside triphosphate (dNTP) triphosphohydrolase SAM domain and HD domain 1 (SAMHD1) was shown to limit the efficacy of ara-C by intracellularly hydrolyzing its active metabolite (PMID: 27991919). Other nucleoside analogs, such as clofarabine, fludarabine, and gemcitabine have also been shown to be substrates of SAMHD1 (PMID:30305425). Among adult AML patients, higher SAMHD1 expression in the leukemic cells has been found to correlate with poor outcome (PMID: 30341277). However, whether genetic variation in SAMHD1 has any impact on the clinical outcomes in pediatric AML patients has not been evaluated in depth. In this study, we looked into 25 single nucleotide polymorphisms (SNPs) within SAMHD1 gene for their association with clinical outcome of newly diagnosed pediatric AML patients in 2 cohorts (multi-site AML02 [NCT00136084, n=167] and AML08 [NCT00703820, n=231] clinical trials). Briefly, genotypes for the 25 SNPs within SAMHD1 genes were obtained from Illumina 2.5 Omni data of AML patients. Association analysis was conducted using logistic regression models comparing minimal residual disease (MRD) positivity after treatment with chemotherapy using an unadjusted model and an adjusted model stratified by initial risk assignment of the patient determined at diagnosis (i.e. low, standard, and high). MRD positivity was defined as one or more leukemic cell per 1000 mononuclear bone-marrow cells. Cox proportional hazard models were used to examine the association of the SNPs with EFS and OS and considered three modes of inheritance. Cox regression models were performed with or without adjusting for provisional risk group assignment at time of diagnosis. Significance levels for association of SNP with clinical outcome were set at p < 0.05.
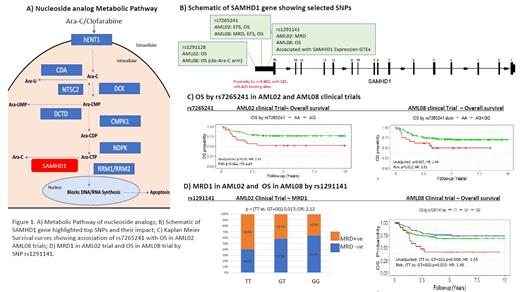
Three top SNPs significantly associated with clinical outcomes were all localized in the 3'UTR region of SAMHD1(Fig 1B). Presence of the variant allele of rs7265241 was associated with a lower EFS and OS in both AML02 and AML08 cohorts. Figure 1C shows the results for OS in AML02 (HR = 2.24, 95% CI (1.05-3.55), p=0.02) and AML08 (HR = 1.52, 95% CI (1.04-1.99), p=0.01). Another 3'UTR SNP, rs1291128 was associated with poor OS in AML02 and in the clofarabine plus ara-C treatment arm in AML08 trial. For rs1291141, T allele was associated with higher MRD positivity (OR: 2.12, p=0.01) in AML02 and poor OS (HR = 1.55, 95% CI (1.12-2.14), p=0.008) in AML08(Fig 1D). Consistent with the clinical outcome results, rs1291141 T allele was also associated with higher SAMHD1 expression in the whole blood (p=8.1e-14) and several other tissues in the GTEx database. https://gtexportal.org/home/
Our results suggest that genetic variation in SAMHD1 is associated with clinical outcomes in pediatric AML patients. Future studies are aimed at looking at haplotypes and SNP-SNP combinations to establish the impact SAMHD1 pharmacogenomics on its expression within leukemic cells and subsequent response to nucleoside analogs in AML patients. SAMHD1 has also been implicated as a tumor suppressor; thus, future studies will evaluate its role both as a proliferation regulator and in drug resistance.
Acknowledgements: NIH-R01-CA132946 (Lamba and Pounds), University of Florida Opportunity Seed Grant (Lamba), American Lebanese Syrian Associated Charities (ALSAC), and American Society of Hematology Bridge funding (Lamba) funded the study. We thank Drs. Campana, Coustan-Smith, and Raimondi for MRD and cytogenetic data.
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal